“Radically different approach” to study of gene expression proposes a unifying model of chromatin regulation and memory

Lacramioara Bintu, photo by Gheorghe Chistol
New research on chromatin regulators offers a unifying model of gene regulation and memory. The work, led by former JCC Fellow Lacramioara Bintu, was published in the February 12, 2016 issue of Science.
Gene expression is regulated in many ways, and in eukaryotes it’s tightly linked to chromatin, the tightly wound DNA structure. Before genetic information from a specific gene can be transcribed, chromatin must modify its shape and allow transcription factors and RNA polymerases to access it. Chromatin regulators, such as specific types of histones and nucleosomes, determine whether or not that happens. Teasing apart the specific mechanisms of gene regulation is key to understanding basic cell biology and disease processes – and realizing its potential for therapeutic applications.
How do chromatin regulators work? How permanent are the changes they induce? How do their effects compare with one another? During her JCC fellowship at Caltech, Bintu focused on these questions. The approach that she and her colleagues took has been called “a radically different approach to dissecting chromatin, focusing not on molecular but on ‘algorithmic’ chromatin biology.”
“Both my postdoctoral advisor, Michael Elowitz, and I have a background in physics and bioengineering,” says Bintu, who will start her own lab at Stanford University in 2017. “So to us it was natural to look at chromatin regulators as devices for controlling gene expression. In this view, you build the device, measure its response, and distill its key characteristics. Therefore, instead of focusing of what chromatin modifications each regulator writes or what proteins it interacts with, we asked: what is the characteristic effect of each chromatin regulator on gene expression and how can we parametrize it?”
Her team engineered cells to express a fluorescent protein, and then tested the action of four chromatin regulators on that gene.
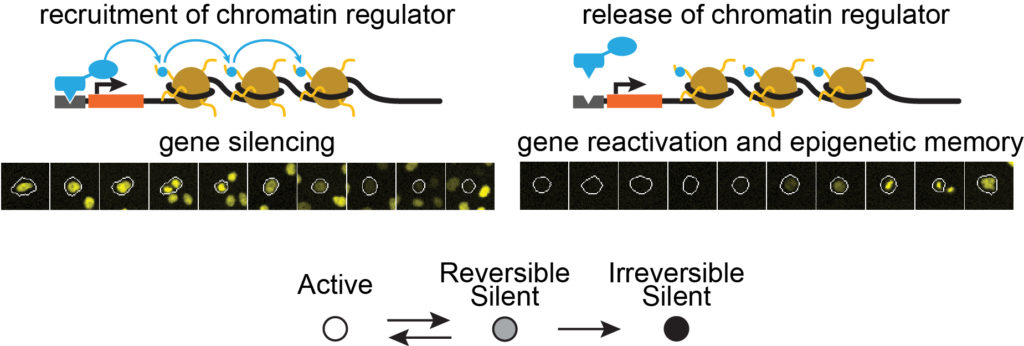
“We really wanted to see changes in gene expression in each individual cell over time,” Bintu explains. “This approach is necessary in order to really get an accurate measurement. For instance, when you see that gene expression was reduced to half, does that mean that all cells have halved their expression or that half of the cells silenced the gene and the other half are still active? It turns out that for chromatin regulators it’s the latter scenario.”
Bintu watched cells grow and divide for 30 days at a time. During that time, she saw cells fluoresce and turn dark, her observations aided by time-microscopy, original cell-tracking software, and flow cytometry. And she found that while each of the chromatin regulators tested had a different effect on gene expression, they could all be summarized in a simple fashion.
“All the chromatin regulators we tested work as digital devices with memory. Recruiting a chromatin repressor changed gene expression in a digital manner, from active to silent. Except that there are two types of silent: reversible and irreversible. From the reversibly silent state, the gene can reactivate. The irreversibly silent state is recorded to memory for a very long time (months). Thinking of chromatin regulation in terms of just three rates—silencing, activation and commitment to an irreversible state—simplifies the system a lot.”
Bintu found that some cells –and their daughter cells– remained dark for days before fluorescing again, long after the regulator disengaged from DNA. Others turned dark only briefly, and still others never recovered.
“The really interesting thing we found is that some of the regulators give this type of hybrid memory where some of the cells awaken while a fraction of the cells remains in a deep coma,” says Bintu. “How many are in the coma depends on how long you gave the signal—how long the chromatin regulator stayed attached.”
Bintu first became interested in gene regulation as an undergraduate at Brandeis University. “When I arrived at Brandeis from Romania, I was sure I was going to study ‘pure’ physics,” she says. “However, I got seduced by a course in Biological Physics taught by Jané Kondev. After the course ended, I continued working with Jané Kondev, Rob Phillips (Caltech) and Hernan Garcia (now at Berkeley) on a thermodynamic model of transcription regulation. While this thermodynamic model describes bacterial transcription well, the packing of DNA into nucleosomes in eukaryotes adds another layer of regulation. I started wondering: ‘What happens when the RNA polymerase encounters a nucleosome?’”
That question became Bintu’s PhD project in Carlos Bustamante’s U.C. Berkeley lab. She used optical tweezers and atomic force microscopy to determine that the polymerase has to wait for the nucleosomal DNA to fluctuate free from histones in order to advance, and that the fate of the histones depends on the speed of the polymerase.
“My last Ph.D. project, examining the direct effect of histone modifications on transcription, convinced me that there are critical questions about chromatin regulation that can only be addressed in the cellular context,” she says. “From my previous work, I was also convinced that taking a quantitative approach and following the time evolution of the process are essential. I decided to join Michael Elowitz at Caltech, who has pioneered dynamic analysis of gene regulation in single cells.”
In January 2017, Bintu will start her own lab in the Bioengineering Department at Stanford University, working to connect the biology of chromatin regulation to the quickly growing field of mammalian cell engineering. “I think that if we have a quantitative, predictive framework for chromatin regulation we can engineer mammalian cells better: for cell therapy and for answering basic biology questions,” she says.
Of her JCC Fellowship, Bintu says she’s very happy that she “became a member of the JCC family.”
“In fact, I first presented a preliminary version of this work at the JCC symposium. In the process, I got really useful feedback both from my JCC mentor, Anjana Rao, and from the audience. This feedback was very useful for follow-up experiments and writing of the manuscript. Moreover, the JCC Foundation was very accommodating of my son, who was only four months old at the time I presented at the JCC Symposium. They encouraged me to bring him and my husband along and gave me a child support stipend. Their attitude, together with the support of the Elowitz Lab, really made me feel that I can succeed in being a good scientist and having a happy family at the same time.”
Abstract figure of the work. Credit: Lacra Bintu and John Yong